Human Coronavirus 2019-nCoV Variants
Variant characterization
SARS-CoV-2 (severe acute respiratory syndrome coronavirus-2), the causative agent of COVID-19, was first reported in Wuhan (China) and has rapidly disseminated around the globe [1]. Only one year after the pandemic outbreak, scientists have developed efficient vaccines and promising therapeutic interventions, such as monoclonal antibodies (mAbs). However, SARS-CoV-2 variants accumulating mutations that confer a selection advantage have emerged [2]. Some are raising concerns as variants might develop resistance to natural immunity and therapies and may jeopardize worldwide control of the disease [3]. The race is on: epidemiologists and immunologists are working hand-in-hand to detect, track, and predict SARS-CoV-2 mutations for adjusted countermeasures.
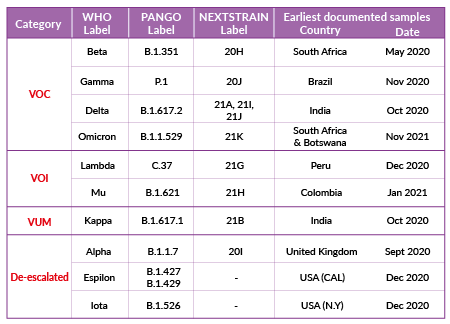
Classification of SARS-CoV-2 variants
SARS-CoV-2's continuous evolution is detected through genome sequencing. SARS-CoV-2 variant classification is based on different labels and is periodically adjusted based on epidemiologic observations. Four labels, proposed by the World Health Organization (WHO) are currently used:
-
Variant of concern (VOC)
A SARS-CoV-2 variant with genetic changes for which there is clear evidence of an increase in transmissibility, disease severity, and/or impact on immune responses and/or drug efficacy.
-
Variant of interest (VOI)
A SARS-CoV-2 variant with genetic changes that are predicted or known to affect transmissibility, disease severity, and/or immune responses, and/or drug efficacy. The evidence is still preliminary or is associated with major uncertainty.
-
Variant under monitoring (VUM)
A SARS-CoV-2 variant with genetic changes that are suspected to affect virus characteristics with some indication that it may pose a future risk, but evidence of phenotypic or epidemiological impact is currently unclear.
-
De-escalated variant
This category comprises formerly monitored variants that have been reclassified based on at least one of the following criteria: (1) the variant is no longer circulating, (2) the variant has been circulating for a long time without any impact on the overall epidemiological situation, (3) scientific evidence demonstrates that the variant is not associated with any concerning properties.
Over the year 2021, emerging variants have been categorized as VUM, VOI, or VOC (e.g. Kappa (B.1.617.1), Lambda (C.37), Mu (B.1.621), Omicron (B.1.1.529)), while some VOC and VOI have been de-escalated (e.g. Alpha* (B.1.1.7), Epsilon (B.1.427)). SARS-CoV-2 variant classification as of December 2021 is summarized in Table 1 (* Alpha is classified as de-escalated according to European Center for Disease Prevention and Control (ECDC), and is still classified as VOC according to WHO).
Note: The terminology of viral variation can be confusing as scientists often use the terms variant, strain, and lineage interchangeably. In a virus phylogenetic tree, a new variant emerges when specific (sets of) mutations are selected. If these mutations produce a virus with distinct phenotypic characteristics, the variant is co-termed a strain. A lineage, or clade, is a phylogenetic cluster of variants associated with an epidemiological event (i.e. rapid geographical spread). The most cited platforms sharing SARS-CoV-2 genomes, GISAID [4], Nextstrain [5], PANGO [6], use distinct nomenclature systems for naming and tracking SARS-CoV-2 variants. To assist with public discussions of variants, experts convened by the WHO [7] have recommended using letters of the Greek alphabet (i.e., Alpha, Beta, Gamma, Delta, etc..). For more clarity, we hereby use the term variant with the WHO nomenclature. The PANGO lineage equivalent is mentioned in brackets.
Mutations of interest in the Spike protein
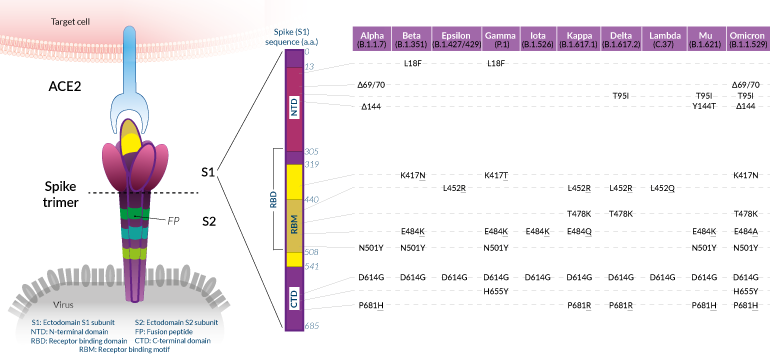
Shared SARS-CoV-2 Spike mutations of interest (click to enlarge)
While mutations have been reported throughout the SARS-CoV-2 genome, many of the variants that have emerged in 2020-2021 share defining amino acid (a.a.) mutations in the Spike (S) protein. Overall, these mutations confer the virus with enhanced ACE2 binding and/or increased antibody evasion [8]. SARS-CoV-2 Spike protein is assembled at the virus membrane as a clove-shaped trimer. Each protomer's ectodomain contains two subunits, S1 and S2. The S1 portion is implicated in viral attachment to the target cell. S1 is comprised of the N-terminal domain (NTD), the ACE2 receptor-binding domain (RBD), and the C-terminal domain (CTD). NTD and RBD are two superantigenic regions, within which mutations have been found to impact both infectivity and immunity. The S2 portion contains elements, such as the fusion peptide (FP), that are critical for viral genome delivery into the target cytosol.
Below, we list mutations in the S1 subunit that have been qualified "of interest" based on the evaluation of their impact on binding to ACE2 and overall antigenicity. Important: this is not an exhaustive list of the a.a. mutations observed in the S protein. We chose to detail and/or illustrate only the mutations that are shared by at least two variants. Reports usually address these mutations, either alone or in combination, in Spike proteins or Spike-pseudotyped viral particles using structural assays (e.g. cryo-electron microscopy), biochemical assays (e.g. ELISA), or cellular assays (e.g. neutralization). To date, mutations in the S2 subunit have not been linked to the virus's spread, nor escape from immunity.
Mutations in Spike RBD:
-
N501Y
The N501Y substitution is prominent in most current variants of concern (Beta, Gamma, Omicron). Position 501 in the S protein is within the receptor-binding motif (RBM) and is one of the six contact amino acid (a.a.) residues with ACE2. N501Y has been shown to improve the binding affinity of SARS-CoV-2 to ACE2 [9, 10].
-
K417N/T
The substitution at K417 to a N (Asparagine) in VOCs Beta and Omicron, or to a T (Threonine) in the VOC Gamma changes a polar a.a. to a neutral one. K417N/T mutations reduce antibody binding at a cost to ACE2 affinity [8, 9].
-
L452R
The substitution at L452 to a R (Purine) in the VOC Delta, VUM Kappa, and de-escalated Epsilon, is located within the RBM [4-7]. While the L452 residue does not directly contact the ACE2 receptor, it has been suggested that the L452R mutation causes structural changes promoting the Spike-Ace2 interaction and escape from neutralizing antibodies [8, 11-13].
-
E484K
The E484K mutation is shared by VOCs Beta and Gamma and the VOI Mu. Like the L452R mutation, the a.a. change at position 484 is located within the RBM and promotes the Spike-Ace2 interaction, as well as escape from neutralizing antibodies [8, 14]. Tests in people vaccinated with the Moderna and Pfizer-BioNTech vaccines suggest E484K and N501Y individually, and both together in combination with K417N, cause a small but significant reduction in neutralization [15]. Omicron, a new VOC (November 2021) also features a substitution at position 484 to a A (Alanine), which impact is yet to be investigated.
Note: Combinations of N501Y, K417N/T, L452R, and E484K/A are found in three of the four current VOCs (Beta, Gamma, Omicron). It is likely that these mutations became rapidly selected owing to their contribution to enhanced SARS-CoV-2 fitness [8, 10].
-
T478K
Like, K452R and E484K, the T478K mutation is is located within the receptor-binding motif (RBM) [4-7]. Less data are available on the effects of this a.a. change, but its increasing global prevalence suggests higher transmissibility and perhaps greater affinity towards the human receptor. Of note, the T478K mutation is shared by the VOCs Delta (and associated clades) and Omicron [4-7]. Further studies are required to determine the overall impact of this mutation.
Mutations in Spike S1-CTD:
-
D614G
Structural studies suggest that the D614G substitution increases the ability to shift the RBD into the "up" position required for ACE2 receptor interaction [9]. Consistent with this, D614G has been shown to increase SARS-CoV-2 infection and transmissibility [10, 11]. This mutation is found in all the variants that have emerged after the original isolate/strain Wuhan-Hu D614 [4-7, 12].
-
H655Y and P681H/R
These substitutions are adjacent to the SARS-CoV-2 furin cleavage site (aka S1/S2 cleavage site) [4-7]. Further studies are required to determine whether they affect furin cleavage efficiency, and therefore increased infection of target cells [10].
Mutations in Spike S1-NTD:
The NTD of the SARS-CoV-2 Spike protein is the least conserved domain [13]. Several substitutions and deletions are found within this region. Notably, 90% of the deletions in Spike occur in four regions within the NTD that are named "recurrent deletion regions" (RDR1 to RDR4) [20]. These RDRs cover external surfaces containing antibody epitopes and therefore contribute to SARS-CoV-2 resistance to neutralizing antibodies [20].
-
L18F
The substitution of L18 to a F (Phenylalanine) is found in VOCs Beta and Gamma [4-7]. The L18F mutation has been predicted to impact mAb binding, based on structural data [13, 21]. Further functional studies are now required to assess this hypothesis.
-
Del69-70
Del69-70 has been observed in the former VOC Alpha as well as in the current VOC Omicron [4-7]. Although this deletion is found in a prominent exterior loop of the Spike, it has not been associated with escape from NTD antibodies, using in vitro neutralization assays with convalescent patients or vaccinee sera [kemp, xie]. Del69-70 is associated with 2-fold increased Spike incorporation into virions but not increased cell-cell fusion. Further studies are required to determine the overall impact of this mutation.
-
Del144/5
The deletion of one of two adjacent Y (Tyrosine) in position 144/145 is another key signature of the former VOC Alpha and current VOC Omicron [4-7]. This mutation is adjacent to glycosylation sites which may play a role in SARS-CoV-2 attachment to target cells or in escaping neutralizing antibodies [13, 21, 24].
SARS-CoV-2 variants of concern
Note: Each SARS-CoV-2 variant listed below is depicted as the phylogenetic root node of a variant clade, which comprises multiple genomic sequences. However, most if not all genomic sequences in that clade share a list of defining mutations. InvivoGen provides one Spike consensus coding sequence for each variant, which can be downloaded from the plasmid product pages.
-
Beta (B.1.351)
The Beta (B.1.351) variant emerged and became the dominant lineage in South Africa in October 2020 [25]. Over the year 2021, Beta has spread to all continents [4-7]. Three S.A strains (B.1.351 v1, B.1.351 v2, and B.1.351 v3) have been reported. They all share the same mutations within the RBD: K417N, E484K, and N501Y, which are thought to increase the virus transmission potential (see above). Each B.1.351 variant carries an additional and unique set of Spike mutations [12]. Some of these mutations have been associated with decreased in vitro neutralization potency by convalescent plasma (~10 fold), and sera from Moderna and Pfizer vaccinees (~5-6 fold) [12, 24]. Other SNPs in the B.1.351 lineage are found in ORF1a, Nucleocapsid, and Envelope [https://covariants.org/variants/20H.Beta.V2].
-
Gamma (P.1)
The Gamma (P.1) variant was detected in December 2020 in the Amazonas state, North Brazil [26, 27] before it started spreading to Japan, North America, Europe, and Australia [4-7]. It shares critical Spike RBD mutations (K417N/T, E484K/A, N501Y) with VOCs Beta and Omicron. In vitro assays have associated K417N/T and E484K substitutions with increased ACE2 binding and SARS-CoV-2 escape from mAbs and convalescent COVID-19 patient sera [8, 9 15] (see above). Other SNPs in the P.1 lineage are found in ORF1ab, ORF8, Nucleocapsid, and Spike (including L18F, T20N, P26S, D138Y, R190S, H655Y, T107I, V1116F) [https://covariants.org/variants/20J.Gamma.V3].
-
Delta (B.1.617.2)
The Delta (B.1.617.2) variant was first detected in India in late 2020 and expanded rapidly, dominating in almost every country by late 2021 [4-7]. Interestingly, it does not share the critical N501Y substitution with VOCs Beta and Gamma, but features the L452R mutation, conferring Delta with enhanced ACE2 affinity, and which thus, may compensate for N501Y absence [8]. Moreover, this variant harbors T478K and P681R, also found in the VOC Omicron, which may increase the virus infectivity (see above). Other SNPs in the B.1.617.2 lineage are found in ORF1b, ORF3a, ORF7a, ORF8, ORF9b, Nucleocapsid, and Spike (including T19R, del156-157, R158G, D950N) [https://covariants.org/variants/21A.Delta].
-
Omicron (B.1.1.529)
The Omicron (B.1.1.529) appears to have arisen in November 2021 in South Africa [4-7]. This variant is primarily of concern as it has spread to all continents within 2 weeks. Moreover, Omicron displays a very large number of mutations, in the Spike sequence as well as elsewhere in the genome [https://covariants.org/variants/21K.Omicron]. Many mutations within the Spike RBD and NTD, which are shared to some extent with other variants, are all combined in the Omicron sequence, including del69-70, del144, K417N, T478K, E484A, N501Y, H655Y, and P681H. Omicron has drawn immediate attention to scientists who are dissecting the impact of its mutational changes on transmissibility and immunity [28].
To help you study the impact of SARS-CoV-2 variants, InvivoGen offers:
➤ Plasmids harboring the genes encoding SARS-CoV-2 Spike. These ready-to-go vectors have been designed for mammalian cell expression or protein production and secretion.
➤ Cell lines derived from the human embryonic kidney 293 (HEK-293) or the human A549 lung carcinoma cell lines. They have been specifically designed to study SARS-CoV-2 infection, as well as for the development of novel therapeutics.
➤ Fusion proteins consisting of Spike fragments with a C-terminal poly-histidine, human IgG1 Fc, or Lucia luciferase tag.
➤ Antibodies targeting the SARS-CoV-2 Spike RBD. These antibodies are available with different immunoglobulin isotypes, either native or engineered.
Products
| Spike (S) and S-derived Coding Sequences | SARS-CoV-2 full Spike, S1, and RBD coding sequences |
| COVID-19-Related Cell Lines | ACE2-(TMPRSS2) expressing cells – Cytokine reporter cells |
| SARS-CoV-2 Spike RBD Proteins | Recombinant fusion proteins produced in CHO cells |
| SARS-CoV-2 Spike S1 Proteins | Recombinant fusion proteins produced in CHO or HEK293 cells |
| COVID-19- Related Antibodies | Recombinant monoclonal antibodies – multiple isotypes |
References
1. Zhu, N. et al. 2020. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med 382, 727-733.
2. Smith, E.C. & Denison, M.R. 2013. Coronaviruses as DNA Wannabes: A New Model for the Regulation of RNA Virus Replication Fidelity. PLOS Pathogens. 9(12):e1003760.
3. Wu, A. et al. 2021. One year of SARS-CoV-2 evolution Cell Host & Microbe. DOI:10.1016/j.chom.2021.02.017.
4. https://www.gisaid.org/
5. https://nextstrain.org/
6. https://cov-lineages.org/
7. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
8. Mannar D. et al., 2021. Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding. Cell Reports. DOI: 10.1016/j.celrep.2021.110156.
9. Starr, T. N. et al., 2020. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 182 (5): 1295–1310.e20.
10. Gong S.Y. et al., 2021. Contribution of single mutations to selected SARS-CoV-2 emerging variants Spike antigenicity. bioRxiv 2021.08.04.455140; DOI: https://doi.org/10.1101/2021.08.04.455140.
11. Deng, X. et al., 2021. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation. medRxiv. DOI:10.1101/2021.03.07.21252647.
12. Garcia-Beltran, W.F. et al., 2021. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 184:1-12. DOI: 10.1016/j.cell.2021.03.013.
13. Plante, J.A. et al., 2021. The variant gambit: COVID-19's next move. Cell Host & Microbe. DOI: 10.1016/j.chom.2021.02.020.
14. Greaney, A.J. et al., 2021. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escapes antibody recognition. Cell Host & Microbe. DOI: 10.1016/j.chom.2020.11.007.
15. Wang Z. et al., 2021. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 592(7855):616-622.
16. Yurkovetskiy, L. et al., 2020. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 183(3):739-751.e8.
17. Plante J.A. et al., 2020. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. DOI: 10.1038/s41586-020-2895-3.
18. Hou, Y.J. et al., 2020. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 370(6523):1464-1468.
19. Korber B. et al., 2020. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases the infectivity of the COVID-19 virus. Cell. 182:1-16.
20. McCarthy, K.R. et al., 2021. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science. DOI: 10.1126/science.abf6950.
21. McCallum, M. et al., 2021. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 184(9):2332-2347.e16.
22. Kemp, S.A. et al., 2021. Recurrent emergence and transmission of a SARS-CoV-2 Spike deletion H69/V70. BioRXiv DOI: 10.1101/2020.12.14.422555.
23. Xie, X. et al., 2021. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat Medicine. DOI: 10.1038/s41591-021-01270-4.
24. Wang P. et al., 2021. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 593:130-135.
25. Tegally, H. et al., 2021. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 592:438-443.
26. Faria, N.R. 2021. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586.
27. Sabino, E.C et al., 2021. Resurgence 821 of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet 397, 452-455.
28. Martin D.P. et al., 2021. Selection analysis identifies significant mutational changes in Omicron that are likely to influence both antibody neutralization and Spike function (parts 1 and 2). https://virological.org/t/selection-analysis-identifies-significant-mutational-changes-in-omicron-that-are-likely-to-influence-both-antibody-neutralization-and-spike-function-part-1-of-2/771 and https://virological.org/t/selection-analysis-identifies-significant-mutational-changes-in-omicron-that-are-likely-to-influence-both-antibody-neutralization-and-spike-function-part-2-of-2/772.